
Application Note
Organoids for disease modeling and in vitro drug screening
- Increase throughput and quantitative assessment of phenotypic changes in organoids with automated imaging and analysis
- Automate workflow for monitoring, maintenance, and characterization of organoids
- Overcome complexity of organoid image analysis using machine learning tools
Oksana Sirenko, PhD | Sr. Scientist | Molecular Devices
Angeline Lim, PhD | Applications Scientist | Molecular Devices
Mary Kassinos | Cell Culture Technician | Molecular Devices
Introduction
3D cell models representing various tissues were successfully used for studying complex biological effects, tissue architecture, and functionality. However, the complexity of 3D models remains a hurdle for wider adoption in research and drug screening.
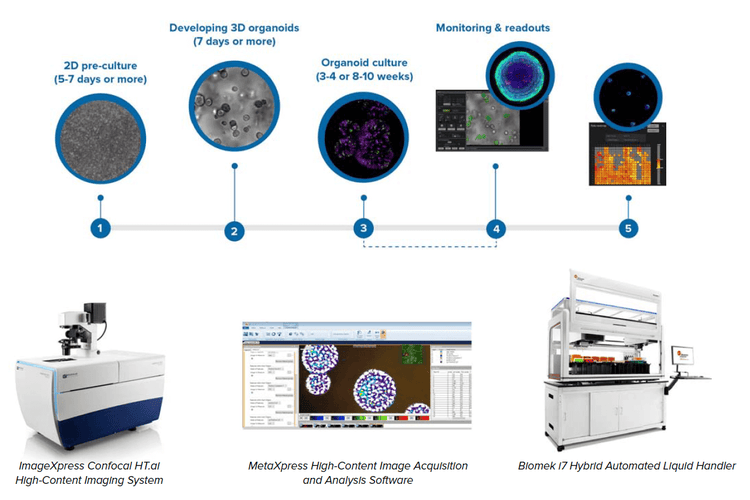
We describe an automated integrated system that would allow automated monitoring, maintenance, and characterization of growth and differentiation of organoids and stem cells, as well as testing the effects of various compounds. The automated, integrated system included an ImageXpress® Confocal HT.ai High-Content Imaging System, automated incubator, Biomek i7 Hybrid workstation, and a robotic device.
The workflow demonstrates the usefulness of automation and advanced high-content imaging for increased throughput and information in organoid assays, critical for compound screening.

Methods
Cell cultures
3D lung organoids: 3D lung organoids were derived from primary human lung epithelial cells (ScienCell). Cells were cultured and expanded in 2D according to ScienCell protocol. For 3D organoid culture, the PneumaCult™ Airway Organoid Kit (STEMCELL Technologies) was used according to manufacturer’s protocol. Briefly, cells were seeded in 90% Matrigel (Corning) domes in 24-well plate format (one dome per well) and were fed every second day for two weeks using the PneumaCult Airway Organoid seeding media. Then differentiation was carried out for another six weeks using the PneumaCult Airway Organoid differentiation media.
Cell monitoring and imaging
Transmitted light (TL) of fluorescent images were acquired on the ImageXpress Confocal HT.ai system (Molecular Devices) using MetaXpress® High-Content Image Acquisition & Analysis Software. For organoids, Z-stack images were acquired with the 4X or 10X objectives using confocal mode. MetaXpress software or IN Carta® Image Analysis Software was used for all analyses.
Results
Culturing and imaging 3D lung organoids
3D lung organoids were developed from primary human lung epithelial cells cultured in Matrigel with a mixture of growth factors. Organoids were monitored using transmitted light, then stained and imaged through Matrigel using automated confocal imaging (Figure 1). Image analysis included conventional and AI-based tools. Developing organoids comprised spherical objects with complex morphology including cavities, protrusions, and vesicle structures. Increases in size and complexity were monitored during 6–8 weeks of development. Advanced image analysis allowed 3D reconstitution and complex analysis of organoids, cell morphology, viability, and differentiation markers. We characterized multiple quantitative descriptors of organoids that could be used for studying disease phenotypes and compound effects. We measured concentration-dependent effects of several drugs that have been known to cause lung toxicity (Figure 2).
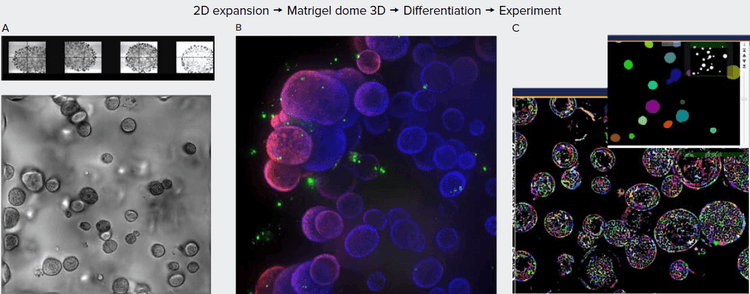
Figure 1. A. Organoids in Matrigel dome after four weeks in culture, TL images (4X, 10X). B. Organoids six weeks in culture stained with Hoechst dye (blue) and MitoTracker (red), 10X. Organoids imaged using confocal option, Z-stack of 23 images 10 μm apart. Maximum projection image presented. C. Image analysis using Custom Module Editor (MetaXpress software). Finding organoids, cells, and subcellular structures.
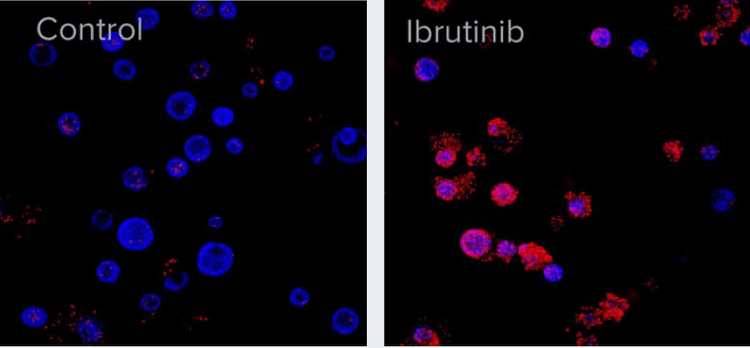
Figure 2. A. After six weeks of development, organoids were treated with 10 μM of Ibrutinib for 72 h. Organoids were then stained with Hoechst nuclear dye (blue) and EtHD-1 (red) to detect dead cells and imaged using ImageXpress Confocal HT.ai system. Numbers of EtHD-1 positive (dead) and negative (live) cells were counted using 3D analysis and used to determine EC50.
The organoid culture was started from primary lung epithelial cells (see Methods section), and then organoids were grown in Matrigel domes using reagents and protocol from STEMCELL Technologies. Briefly, cells were first expanded in 2D, then mixed with GF-reduced Matrigel and seeded into Matrigel domes in 24-well or 96-well plate formats (Figure 1).
Automation of cell culture and imaging protocols
Organoids are an essential tool for disease modeling and assessment of compound effects. Automated imaging and analysis of organoids are important for quantitative assessment of phenotypic changes in organoids, and for increasing the throughput for experiments and tests.
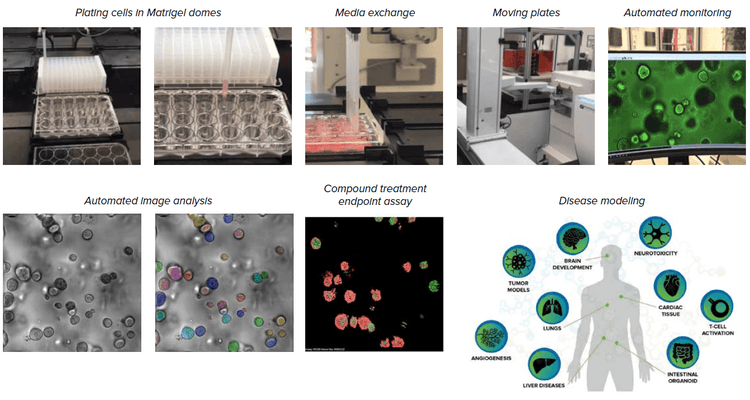
We built an automated, integrated system that would allow automated monitoring, maintenance, and characterization of growth and differentiation of organoids and stem cells, as well as testing the effects of various compounds. The automated, integrated system includes ImageXpress Confocal HT.ai system and analysis software, automated CO2 incubator, Biomek i7 liquid handler, collaborative robot and rail, as well as additional optional instruments (automated centrifuge, ImageXpress® Pico system, plate reader).
Components of the automated organoid workflow
Conclusion
- The process for organoid development can be automated by integrating several instruments, providing automated cell culture, maintenance, and differentiation of 3D cellular models that can be used for compound screening a variety of assays.
- Cell cultures can be monitored in transmitted light with the AI-based image analysis that allows the detection and characterization of organoids.
- Confocal imaging in combination with 3D analysis allows complex, quantitative analysis of cellular content of organoids as well as count and measurements of cells with different phenotypes (cell count, live-dead assessment, cell scoring for specific markers, etc.). The methods can be used for testing the effects of different compounds, toxicity evaluation, and disease modeling.