
Application Note
Measure Calcium Oscillation in iCell Cardiomyocytes2 on the FLIPR Tetra System
- Improve assay workflow and quality with shorter cell culture time
- Identify compounds that could cause safety issues earlier in the drug discovery process
- Simplify data analysis with ScreenWorks Peak Pro Software
Carole Crittenden | Applications Scientist | Molecular Devices
Oksana Sirenko, PhD | Research Scientist | Molecular Devices
Introduction
Costs of screening for efficacy and safety of compounds continue to increase which creates a need for innovative technologies to improve characterization earlier in the drug discovery process. The FDA is developing guidelines for testing compounds to help ensure drug safety so that drugs will not need to be pulled from the market due to off-target effects such as blocking hERG channels in the heart and causing symptoms like Torsades de pointes. This effort is called the Comprehensive in vitro Proarrhythmia Assay or CiPA Project.
Induced pluripotent stem cell (iPSC)- derived cardiomyocytes are a particularly attractive in vitro model system due to their use for evaluation of compound effects on both cardiac function and safety. Calcium signal oscillation reflects changes in cytoplasmic calcium concentration making it possible to use a calcium sensitive dye such as the EarlyTox™ Cardiotoxicity Kit. ScreenWorks® Peak Pro Software analyzes peak frequencies, peak width at 10% amplitude, and other parameters to characterize compound effects on cardiomyocytes in a high throughput screening format.
Cellular Dynamics International has developed an enhanced version of their iPSC-derived cardiomyocytes that recovers from being frozen faster than previous version of iPSCs. This development provides an improvement in the workflow by shortening the growth timeline by 5-7 days when screening compound effects on calcium transients in beating cardiomyocytes on the FLIPR® Tetra High-Throughput Cellular Screening System. A sample workflow is shown in Figure 1. Here, we describe the testing of cancer compounds some of which are known to interfere with hERG channels and other compounds known to block hERG.
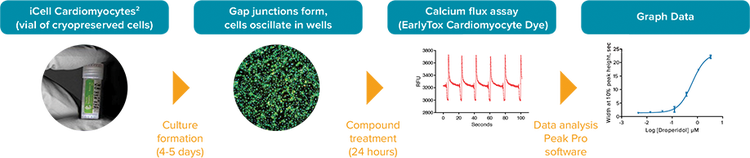
Figure 1. iPSC2 cardiomyocyte assay workflow using the FLIPR Tetra System to measure change in calcium oscillation.
Materials
- iCell® Cardiomyocytes 2 Kit including cells, plating medium, and maintenance medium (Provided by Cellular Dynamics,International)
- 0.1% (w/v) gelatin
- 384-well black, clear bottom plate(Corning PN 3712)
- EarlyTox Cardiotoxicity Kit(Molecular Devices PN R8210)
- Test compounds and DMSO(Sigma Aldrich)
- FLIPR Tetra High-Throughput Cellular Screening System (Molecular Devices)
Prepare the plate using iCell Cardiomyocytes2
384-well, black, clear bottom culture plates were prepared by coating with 0.1% gelatin following the instructions in the iCell Cardiomyocytes2 User Guide. The iCell Cardiomyocytes2 were thawed and plated at 8,000 cells/well in 25 µL plating medium. Cells were incubated at 37°C, 5% CO2 in 100% humidity for 24 hours. The cells formed a monolayer and the plating media was exchanged with maintenance media. The cells were fed with maintenance media every other day. On the fifth day, the cells were oscillating together and ready to use.
Prepare the plate for screening
For screening, compound stock solutions were prepared at 10 mM in 100% DMSO. Test solutions were prepared at 10-100 µM in maintenance media. The solutions were added to the wells for 24 hours prior to reading on the FLIPR Tetra System. The final concentration of DMSO in media was 0.15% (v/v). The compounds were tested in either duplicate (n=2) or triplicate (n=3) at concentrations ranging from 30 or 100 µM with half-log dilutions.
Observe calcium oscillations using the FLIPR Tetra System
24 hours prior to assay, half log dilutions of various compounds starting with a final concentration of 30 µM were added to the iPSC Cardiomyocytes2 with a media change. Two hours prior to assay, 25 µL of 2X concentration of the EarlyTox Cardiotoxicity dye in maintenance media was added to each well and incubated at 37°C, 5% CO2. Changes in fluorescence due to calcium concentration as the cells oscillated were read on the FLIPR Tetra System immediately after removal from the incubator. Setup parameters used in the ScreenWorks software read protocol are shown in Table 1. The data was analyzed using the ScreenWorks Peak Pro Software module and graphs were made in GraphPad Prism 6.
Table 1. FLIPR Tetra System read parameters
Compound effects on cardiomyocyte oscillation peak parameters
Calcium oscillations at 24 hours after compound addition were read using the FLIPR Tetra System. iPSC2s performed in the same manner as previous versions of iPSCs by forming gap junctions and oscillating together to give peak frequencies measured with calcium sensitive dye. Cells were ready to assay 4-5 days earlier than previous versions of the cells which showed an improvement to the screening workflow due to a lower risk of contamination and an overall shorter timeframe while producing similar results.
The compounds studied included anticancer compounds: Sunitinib, Imatinib and Staurosporine (not a clinical compound due to toxicity) which are kinase inhibitors; Busulfan, an alkyl sulfonate; Doxorubicin an anthracycline; Etoposide, a topoisomerase II inhibitor; and Paclitaxel which interferes with cellular tubulin production. At 24 hours post addition of compound, some calcium oscillation patterns varied widely from the control pattern (Figure 2). Sunitinib had a slower rate of oscillation in addition to a much larger peak width at 10% of peak height compared to the control. Other anticancer compounds with similar patterns included Imatinib and Staurosporine. These three compounds have been associated with blocking hERG channels. hERG represents a group of genes that code for potassium channels that contribute to the recovery of the heart beat and can contribute to long Q-T syndrome or Torsades de pointes syndrome. Paclitaxel, Busulfan, and Etoposide are not known to have hERG activity and did not have the same slowing of calcium oscillation nor did they have wider peaks at 10% of peak height. The concentration response curves demonstrated a decrease in peak frequency after 24-hour exposure to Doxorubicin and Sunitinib. Paclitaxel was also an example of a compound that did not affect peak frequency (Figure 3).
In addition to anticancer compounds, known hERG blocking compounds Droperidol, an antipsychotic and dopamine receptor blocker; Amitriptyline, a tricyclic antidepressant and serotonin receptor inhibitor; and Cisapride, an antacid, all known for long Q-T interval prolongation were also tested in the assay. When added to the iPSC2s for 24 hours, all three compounds exhibited a slowing in peak frequency and an increase in peak width at 10% peak amplitude (Figure 4). A summary of IC50 values of all compounds tested in the calcium oscillation assay is shown in Table 2.
Peak Frequency IC
50
(µM)
Table 2. Compound effects on calcium oscillation in iPSC2 cardiomyocytes.
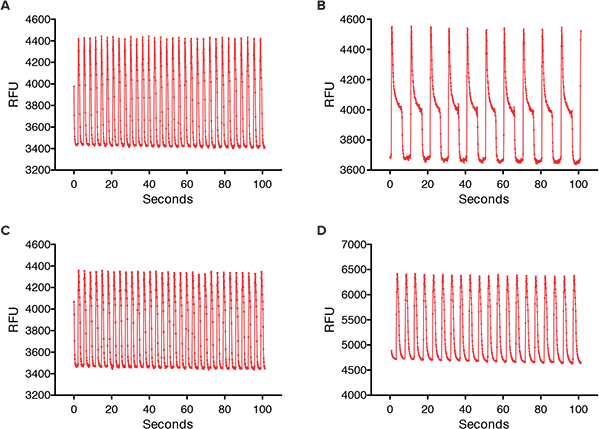
Figure 2. Calcium oscillations in iPSC2 cardiomyocytes. (A) Control calcium oscillations measured on the FLIPR Tetra System. (B) Calcium oscillations after 24 hour exposure of iPSC2 cells to 1 µM Sunitinib.(C) Calcium oscillations after 24 hour exposure of iPSC2 cells to 1 µM Paclitaxel. (D) Calcium oscillations after 24 hour exposure of iPSC2 cells to 1 µM Doxorubicin.
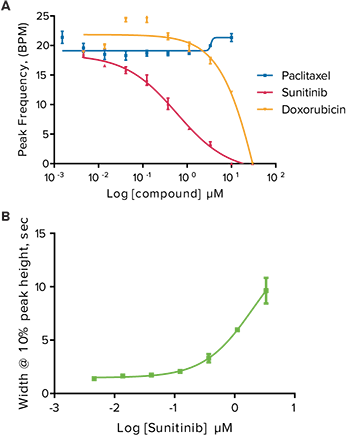
Figure 3. Anticancer compound effects on peak parameters in oscillating cardiomyocytes. (A) Peak frequency (BPM) of calcium oscillations in iPSC2 cells after 24-hour exposure to increasing concentrations of Sunitinib, Doxorubicin, and Paclitaxel. Note that Paclitaxel did not slow the peak frequency. (B) Increase in peak width at 10% peak height with increasing concentration of Sunitinib.
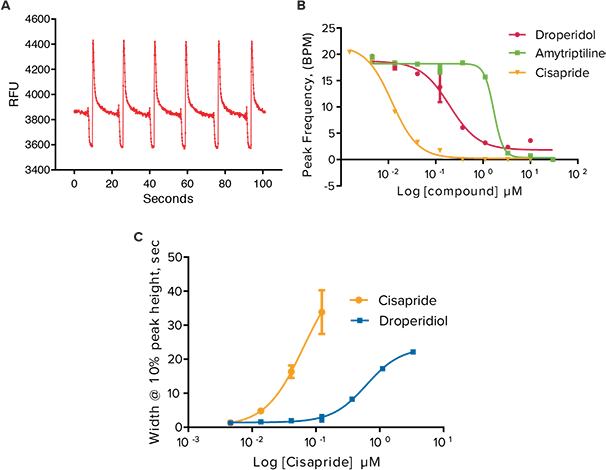
***Figure 4. Effects of selected hERG blockers on peak parameters after 24 hour exposure in oscillating iPSC cardiomyocytes. (A)**Calcium oscillation pattern after exposure to 1 µM Cisapride. **(B)*Peak frequencies after exposure to Droperidol, Amytriptiline, and Cisapride. (C) Increase in peak width at 10% peak height.
Conclusion
iPSC2 cardiomyocytes improved the screening assay workflow by allowing calcium oscillation assays on the FLIPR Tetra System to be monitored at 4-5 days rather than 10-14 days post thawing of cells. ScreenWorks Peak Pro Software easily analyzed oscillation patterns to demonstrate compound effect on cardiomyocytes and was an efficient tool for the drug screening process. This assay can be performed early in the process to help identify compounds with potentially adverse or beneficial effects on the heart before they are brought forward into the lead discovery process.
Resources
- Sirenko, Oksana, et al. “Phenotypic Assays for Characterizing Compound Effects on Induced Pluripotent Stem Cell-Derived Cardiac Spheroids.” ASSAY and Drug Development Technologies, vol. 15, no. 6, 2017, pp. 280–296., doi:10.1089/adt.2017.792.
- Sirenko, Oksana, et al. “In vitro cardiotoxicity assessment of environmental chemicals using an organotypic human induced pluripotent stem cell-Derived model.” Toxicology and Applied Pharmacology, vol. 322, 2017, pp. 60–74., doi:10.1016/j.taap.2017.02.020.
- Sirenko, Oksana, et al. “Assessment of beating parameters in human induced pluripotent stem cells enables quantitative in vitro screening for cardiotoxicity.” Toxicology and Applied Pharmacology, vol. 273, no. 3, 2013, pp. 500–507., doi:10.1016/j.taap.2013.09.017
- “iCell Cardiomyocytes 2 User Guide.” Cellular Dynamics International, Aug 2016