
Application Note
IMAP phosphodiesterase assays on SpectraMax Multi-Mode Microplate Readers
- Non-antibody-based platform
- Sensitive and stable
- Complete, homogeneous assay system
- FP and TR-FRET options
Introduction
IMAP®Technology from Molecular Devices enables rapid, homogeneous, and non-radioactive assay of kinases, phosphatases, and phosphodiesterases and is suited for both assay development and high-throughput screening. IMAP assaysare based on binding of phosphate to immobilized metal coordination complexes on nanoparticles. When IMAP binding entities bind to phosphorylated substrate, molecular motion of the peptide is altered, and fluorescence polarization (FP) for the fluorescent label attached to the peptide increases (Figure 1, left). In a TR-FRET version of the assay, the inclusion of a Terbium (Tb) donor enables a fluorescent energy transfer to occur when phosphorylated substrate is present (Figure 1, right). This assay is detected in a time-resolved mode, which virtually eliminates fluorescence interference from assay components or compounds in a screen. TR-FRET also offers flexibility in substrate size and concentration.
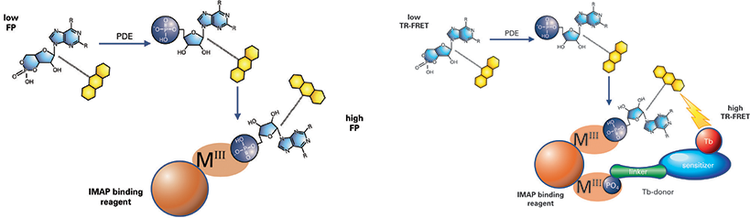
***Figure 1. IMAP FP and TR-FRET phosphodiesterase assay principle.*A phosphodiesterase reaction is performed using a fluorescent-labeled substrate. Binding Solution containing large M(III)-based nanoparticles is then added. In the FP readout (left), the small phosphorylated fluorescent substrate binds to the large nanoparticles, which reduces the rotational speed of the substrate and thus increases its fluorescence polarization. In the TR-FRET readout (right), phosphorylated substrate and Tb donor both bind to the nanoparticle, bringing the Tb donor in close proximity to the fluorescein acceptor on the substrate and enabling FRET.
Cyclic nucleotide phosphodiesterases (PDEs) are a group of enzymes that degrade the phosphodiester bond of cAMP and cGMP, second messengers that are involved in a variety of biological processes. They have emerged as a key class of druggable targets due to their clinical significance in areas including heart disease, dementia, depression, and others. In this application we demonstrate how PDE enzyme dilution and inhibition curves are performed with IMAP Technology using the SpectraMax®i3, SpectraMax®Paradigm®, SpectraMax®M5, and FlexStation®3 Multi-Mode Microplate Readers with SoftMax®Pro Software.
Materials
- IMAP FP PDE Evaluation Kit (Molecular Devices P/N R8175)
- IMAP TR-FRET PDE Evaluation Kit (Molecular Devices P/N R8176)
- IMAP Progressive Binding Reagent
- IMAP Progressive Binding Buffer A (5X)
- IMAP Progressive Binding Buffer B (5X)
- TR-FRET Tb Donor
- IMAP Reaction Buffer with 0.01% Tween-20 (5X)
- Fluorescein-labeled cGMP Substrate (Molecular Devices P/N R7507)
- Phosphodiesterase 1, 3’, 5’-cyclicnucleotide-specific from bovine brain (Sigma P/N P9529)
- Calmodulin (Sigma P/N P1431)
- Calcium chloride (major laboratory suppliers)
- DL-Dithiothreitol (DTT) (major laboratory suppliers)
- 384-well polystyrene black (for FP) and white (for TR-FRET) microplates (e.g., Corning P/N 3573 and 3572)
Methods
Enzyme dilution series
Complete reaction buffer (CRB) was prepared with 1 mM DTT, 2500 U/mL Calmodulin, and 2.5 mM CaCl2. Fluorescein-labeled cGMP substrate was diluted in CRB to make a 200 nM (2X) working concentration. Optimal substrate concentration and other assay parameters were determined previously (IMAP Assay Archive). A 1:3 dilution series of PDE1 enzyme, starting at 0.002 U/mL, was prepared as 2X working dilutions in CRB. Enzyme reactions were assembled in quadruplicate at a volume of 20 μL per well in a black (FP) or white (TR-FRET) 384-well plate with the following additions:
- 10 μL of 2X enzyme dilution or no-enzyme (CRB)
- 10 μL of 2X substrate solution
For buffer-only controls, 20 μL CRB was added to wells. Assays were incubated at room temperature for one hour.
Inhibitor assays
The PDE1 inhibitors 8-MM-IBMX and zaprinast were serially diluted in CRB at 4X working concentrations. Both series started at 200 μM (final in assay) and were diluted 1:3. PDE1 enzyme with EC80concentration of 1.3 x 10-4U/mL was prepared as a 4X working solution. Reactions were assembled as follows:
- 5 μL of 4X inhibitor or CRB for controls
- 5 μL of 4X enzyme
- 10 μL of 2X substrate solution
Assays were incubated at room temperature for one hour.
Binding reaction
IMAP Binding Solution specific to each assay detection mode was prepared. For FP, Binding Solution contained 75% Buffer A, 25% Buffer B, and 1:600 Binding Reagent. For TR-FRET, it contained 60% Buffer A, 40% Buffer B, 1:800 Binding Reagent, and 1:400 Tb Donor. Buffer-only controls received Binding Solution minus Tb Donor. 60 μL of the appropriate Binding Solution was added to each assay well, and assays were incubated for one hour (FP) or overnight (TR-FRET).
Detection
At the completion of the binding step, assay plates were read on the SpectraMax i3 and SpectraMax Paradigmreaders using the settings in Table 1. All data were acquired and analyzed using SoftMax Pro Software. Preconfigured software protocols containing optimized settings and analysis parameters for each assay were used to simplify calculation of results. The Read Height Adjustment procedure was run prior to reading the plate to ensure optimal read height for each assay.
FP-FLUO
Ex 485 nm/Em 535 nm
TR-FRET B/G (custom)
Wavelength 1: Ex 340 nm/Em 490 nm
Wavelength 2: Ex 340 nm/Em 520 nm
Integration time: 100 ms
Read from Top
Read height 4.27 mm
Integration time: 1 ms
Excitation time: 0.05 ms
Number of pulses: 100
Meas. Delay: 0.2 ms
Read from Top
Read height 6.02 mm
*This plate definition in SoftMax Pro Software is the same for black and white plates.
Table 1. SpectraMax i3 reader settings for IMAP PDE assays.
Results
Performing an enzyme dilution series verifies the enzyme and assay performance and allows calculation of the EC80concentration of enzyme that will be used for inhibitor screening assays. PDE1 enzyme dilution series were performed using IMAP Technology in both FP and TR-FRET assay modes. EC50values agreed very closely for both assay types (Figure 2) and were similar to previously published values. The Z’ factor calculated from positive and negative controls was greater than 0.9 for each assay, indicating robustperformance suitable for high-throughput screening. Inhibitor assays yielded similar IC50 values for both FP and TR-FRET assay readouts (Figure 3).
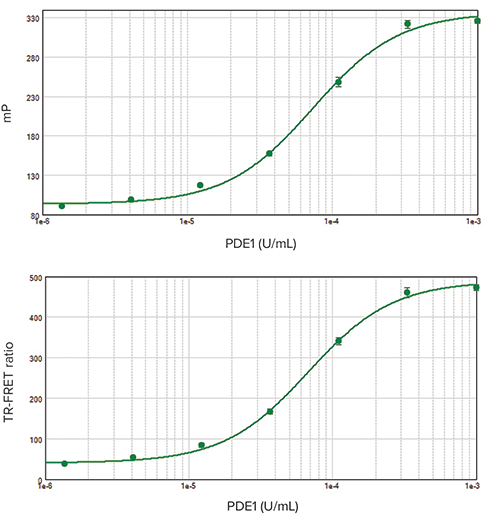
***Figure 2. EC50curves for IMAP FP and TR-FRET readouts. Top:**FP; EC50value was 7.3 x 10-5 units/mL with a Z’ factor of 0.95. **Bottom:*TR-FRET; EC50value was 6.8 x 10-5units/mL with a Z’ factor of 0.92.
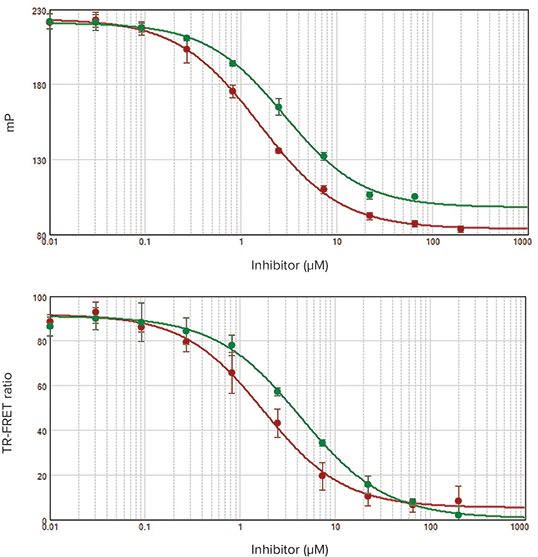
***Figure 3. IC50curves for IMAP FP and TR-FRET readouts. Top:*FP. Bottom: TR-FRET. On each plot, IC50curves for IBMX are in red and zaprinast are in green. For 8-MM-IBMX, IC50values of 1.55 µM (FP) and 1.77 µM (TR-FRET) were obtained. For zaprinast, IC50values were 2.83 µM (FP) and 4.35 µM (TR-FRET). These IC50values are consistent with previously published results1.
Conclusion
IMAP Technology offers a homogeneous,non-antibody-based platform for screening kinases, phosphatases, and phosphodiesterases. Stable and robust, it is amenable to miniaturization and yields high-quality data with excellent Z’ factor values, making it suitable for inhibitor screening. Unlike some other methods, IMAP measures direct binding for more relevant results. IMAP TR-FRET assays also enable direct Km determination.
IMAP assayscan be performed on systems shown on the next page. Data presented here were generated with the SpectraMax i3 reader. Similar results were obtained with the SpectraMax Paradigm reader, which offers the added advantage of fast, dual-PMT detection. SoftMax Pro Software with preconfigured assay protocols simplifies the workflow, from data acquisition to analysis. Molecular Devices offers a complete IMAP screening solution, including an IMAP Assay Archive with reference data on over 120 different enzymes, validated protocols, kits, and substrates2.
References
- Ahn, HS, Bercovici, A, Boykow, G, et al. (1997). Potent tetracyclic guanine inhibitors of PDE1 and PDE5 cyclic guanosine monophosphate phosphodiesterases with oral antihypertensive activity. J. Med Chem. 40, 2196-2210.
- IMAP Assay Archive, http://support.moleculardevices.com/assayarchive/index.html.
800 dp
*
20 nmol
120 nmol
R7505
R7506
20 nmol
120 nmol
R7507
R7508
*Data points
Compatible with these Molecular Devices systems

简介
Molecular Devices 的 IMAP® 技术能对激 酶,磷酸酶和磷酸二酯酶进行快速,均质 和非放射性的分析,非常适用于分析开发 和高通量筛选。IMAP 检测是基于磷酸基 团与固定在纳米微粒表面的三价金属阳离 子之间的高亲和力结合。可以使用 FP 与 TR - FRET 两种读数方法来检测磷酸化底 物与纳米微粒相互的结合情况。在 FP 方法 中,当 IMAP 结合溶液与磷酸化底物结合 时,肽的分子运动被改变,引起标记在肽 上的荧光分子的荧光偏振效应 (FP) 增加 ( 图 1,左 )。在 TR - FRET 方法中,当存在 磷酸化的荧光底物时,铽 (Tb) 供体能够与 底物荧光受体发生荧光能量转移 ( 图 1, 右 )。该方法使用时间分辨荧光 (TRF) 检测 模式,能够消除了测定组分或筛选中化合 物的荧光干扰,并且底物尺寸和浓度的选 择更灵活。

***图 1 IMAP FP 和 TR - FRET 磷酸二酯酶检测原理。*使用荧光标记的底物进行磷酸二酯酶反应,然后加入含有大分子 M (III) 纳米颗粒的结合溶液。 在 FP 读数 ( 左 ) 中,小磷酸化荧光底物与大纳米颗粒结合,这降低了底物的旋转速度并因此增加了其荧光偏振。 在 TR - FRET 读数 ( 右 ) 中,磷酸化底物和 Tb 供体均与纳米颗粒结合,使 Tb 供体与底物上的荧光素受体紧密接近,进而产生 FRET 效应。
环核苷酸磷酸二酯酶 (PDEs) 是一组可降解 cAMP 和 cGMP 的磷酸二酯键的酶,是参 与各种生物过程的第二信使。 由于它们在 心脏病,痴呆症,抑郁症等领域具有很现 实的临床意义,已成为一类药物治疗的靶 标分子。 在本应用文章中,我们使用 IMAP 技术进行 PDE 酶稀释实验和生成抑制曲线,实验能在 Spectr SpectraMax®Paradigm®, SpectraMax®M 5 和 FlexStation®3 多功能 读板机上完成,结合 SoftMax®Pro 软件进 行获取和分析数据。
材料
- IMAP FP PDE 评估试剂盒 (Molecular Devices P/N R 8175)
- IMAP TR - FRET PDE 评估试剂盒 (Molecular Devices P/N R 8176
- IMAP 结合试剂
- IMAP 结合缓冲液 A (5x)
- IMAP 结合缓冲液 B (5x)
- TR - FRET Tb 供体
- IMAP 反应缓冲液含 0.01 % 吐温 - 20 (5X)
- 荧光素标记的 cGMP 底物 (Molecular Devices P/N R 7507)
- 牛脑特异性磷酸二酯酶 1,3’,5’- 环 核苷酸 (Sigma P/N P 9529)
- 钙调蛋白 (Sigma P/N P 1431)
- 氯化钙 ( 主要实验室供应商 )
- DL - 二硫苏糖醇 (DTT) ( 主要实验室供应商 )
- 384 孔聚苯乙烯黑色微孔板 ( 用于 FP ) 和白色微孔板 ( 用于 TR - FRET ) ( 例如, 康宁 P/N 3573 和 3572 )
方法
酶稀释
1 mM DTT,2500 U/mL 钙调蛋白和 2.5 mM 氯化钙制备成完全反应缓冲液 (CRB)。 荧光素标记的 cGMP 底物以 CRB 稀释成浓 度为 200 nM (2 X) 的工作液。先前确定了 最佳底物浓度和其它检测参数 (IMAP Assay Archive)。起始浓度为 0.002 U/mL 的 PDE 1 酶用 CRB 按 1∶3 的比例稀释,制成 2 x 的酶稀释液。在黑色 (FP) 或白色 (TR - FRET) 的 384 孔板中加入以下溶液,每孔 总体积 20 μL:
- 10 μL 2 X 酶稀释液或无酶 (CRB)
- 10 μL 2 X 底物溶液
向孔中加入 20 μLCRB,作为空白对照。然 后将检测板放于在室温下,酶反应 1 小时。
抑制剂检测
PDE 1 抑制剂 8 - MM - IBMX 和扎普司特用 CRB 连续稀释,制成 4 x 工作溶液。 两个抑制剂均以初始浓度 200 μM 进行 1∶3 的连续稀释。 制备 EC80 浓度为 1.3×10- 4 U / mL 的 PDE 1 酶作为 4 X 工作溶液。 将以下体积的反应溶液加入微孔板:
- 5 μL 的 4 X 抑制剂或 CRB 用于对照
- 5 μL 的 4 X 酶
- 10 μL 的 2 X 底物溶液
然后在室温下温育 1 小时。
结合反应
制备不同检测方法使用的 IMAP 结合溶液。 FP 检测:结合溶液包括 75 % 缓冲液 A, 25 % 缓冲液 B 和 1∶600 结合试剂。 TR - FRET 检测:结合溶液包括 60 % 缓冲 液 A,40 % 缓冲液 B,1∶800 结合试剂和 1∶400 Tb 供体。不包含 Tb 供体的结合溶 液作为空白对照。 微孔板每孔加入 60 μL 适当的结合溶液, 然后温育 1 小时 (FP) 或过夜 (TR - FRET)。
检测
结合反应完成后,把微孔板放入 SpectraMax i3 和 SpectraMax Paradigm 多功能 微孔读板机中,按照表 1 中的参数设置读 取检测板,然后 SoftMax Pro 软件获取和 分析所有数据。也可以使用软件中内置的 FP 和 TR - FRET 分析方案来优化您的实验 设置和分析参数,简化实验结果的计算。 如果想得到更高精度的读数,在读取板之 前运行读取高度优化程序,确保每次检测 的最佳读取高度。
FP-FLUO
Ex 485 nm/Em 535 nm
TTR-FRET B/G ( 定制 )
波长 1: Ex 340 nm/Em 490 nm
波长 2: Ex 340 nm/Em 520 nm
累计时间:100 毫秒
顶部读数
读取高度 4.27 毫米
累计时间:1 毫秒
激发时间:0.05 毫秒
脉冲数:100
检测延迟:0.2 毫秒
顶部读数
读取高度6.02 毫米
*SoftMax Pro 软件中的黑板和白板的选项是相同的
表 1 用于 IMAP PDE 检测的 SpectraMax i3 酶标仪的参数设置。
结果
酶稀释检测验证了酶的活性和实验灵敏 度,获得的酶 EC80 浓度用于抑制剂筛选检 测。 分别在 FP 和 TR - FRET 检测模式中使用 IMAP 技术进行 PDE 1 酶稀释。两种检测 方法的 EC50 值非常接近 ( 图 2 ) 并且与之前 公布的数值一致。 在每个检测方法中,由阳性和阴性对照计 算的 Z'因子均大于 0.9,这表明 IMAP 技术 适合于高通量筛选的稳健性能。在 FP 和 TR - FRET 读数方法中,两种抑制剂检测获 得了相似的 IC50 值 ( 图 3 )。

***图 2 IMAP FP 和 TR - FRET 读数的 EC50 曲线。*上图:FP; EC50 值为 7.3×10-5 单位/ mL,Z'因子为 0.95。下图:TR - FRET; EC50 值为 6.8×10-5 单位/ mL,Z' 因子为 0.92

图 3 IMAP FP 和 TR - FRET 读数的 IC50 曲线。上图:FP,下图:TR - FRET。。在每个图上,IBMX 的 IC50 曲线为红色,扎普司特曲线为绿色。8 - MM - IBMX:IC50 值为 1.55 μM (FP) 和 1.77 μM (TR - FRET)。Zaprinast:IC50 值为 2.83 μM (FP) 和 4.35 μM (TR - FRET)。这些 IC50 值与先前文献中公布 的结果一致
结论
IMAP 技术提供了一种均相的,非抗体的检 测平台,用于筛选激酶,磷酸酶和磷酸二 酯酶。稳定的荧光信号,可靠的高质量数 据,出色的 Z'因子,这些特点表明 IMAP 技 术非常适用于高通量的抑制剂筛选。与其 它检测方法不同,IMAP 检测采用直接结 合,得到更相关的结果。IMAP TR - FRET 检测还可以直接测定 Km。
本文章中的数据是由 SpectraMax i3 多功 能 微 孔 读 板 机 生 成 的 。 S p e c t r a M a x Paradigm 获得了相似的结果,除此之外, 它还具备快速双 PMT 检测的附加优势。 SoftMax Pro 软件数据库中内置的分析方 案,简化了从数据采集到分析的工作流 程。Molecular Devices 提供完整的 IMAP 技术筛选解决方案,如 IMAP 实验方案,包 括 120 多种不同酶的参考数据,经过验证 的实验方案以及试剂盒和底物。
参考文献
- Ahn, HS, Bercovici, A, Boykow, G, et al. (1997). Potent tetracyclic guanine inhibitors of PDE1 and PDE5 cyclic guanosine monophosphate phosphodiesterases with oral antihypertensive activity. J. Med Chem. 40, 2196-2210.
- IMAP Assay Archive, http://support.moleculardevices.com/assayarchive/index.html.
800 dp
*
20 nmol
120 nmol
R7505
R7506
20 nmol
120 nmol
R7507
R7508
*Data points
Compatible with these Molecular Devices systems
